A Comprehensive Guide to UNet Implementation with TensorFlow
After writing article on Image Segmentation by UNet: Everything you need to know about Implementing in PyTorch, one of my friend asked me…
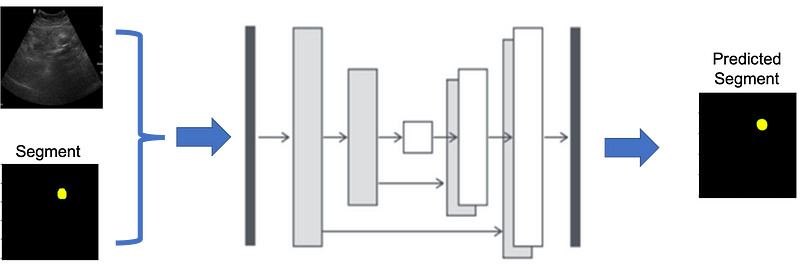
After writing article on Image Segmentation by UNet: Everything you need to know about Implementing in PyTorch, one of my friend asked me to write the same in Tensorflow. So here is the plan of attack to do this:
- Introduction
- Architecture
- Key Features
- Hands on using Tensorflow
- Reference
Introduction
UNet is a type of convolutional neural network (CNN) that was originally developed for biomedical image segmentation. The architecture was introduced by Olaf Ronneberger, Philipp Fischer, and Thomas Brox in their paper “U-Net: Convolutional Networks for Biomedical Image Segmentation,” published in 2015 at the MICCAI (Medical Image Computing and Computer-Assisted Intervention) conference. UNet has since become widely popular in various image segmentation tasks beyond biomedical imaging due to its effective structure for precise localization and the ability to work with a limited amount of training data.
Architecture
The UNet architecture is distinguished by its U-shaped structure, which consists of two main parts: the contraction (downsampling) path and the expansion (upsampling) path.
- Contraction Path (or Encoder): The contraction path follows the typical architecture of a convolutional network. It consists of repeated application of two 3x3 convolutions (unpadded convolutions), each followed by a rectified linear unit (ReLU) and a 2x2 max pooling operation with stride 2 for downsampling. At each downsampling step, the number of feature channels is doubled.
- Expansion Path (or Decoder): The expansion path includes a series of upsampling and convolution steps. Upsampling in the network is achieved through transposed convolutions, which increase the resolution of the feature maps. After each upsampling step, the feature map is concatenated with the correspondingly cropped feature map from the contraction path. This is followed by two 3x3 convolutions, each followed by a ReLU. The concatenation with the high-resolution features from the contraction path allows the network to propagate context information to higher resolution layers, enabling precise localization.
- Final Layer: At the final layer, a 1x1 convolution is used to map the feature vector to the desired number of classes.
Key Features
- Symmetry: The U-shaped architecture ensures that the network has a symmetrical structure, with a downsampling path to capture context and an upsampling path to enable precise localization.
- Skip Connections: The use of skip connections, where feature maps from the downsampling path are concatenated with the feature maps from the upsampling path, helps the network to recover fine-grained details lost during downsampling.
- Efficiency with Data: UNet is designed to work well even with a small number of training images, making it suitable for medical imaging applications where annotated images can be scarce.
Hands on using Tensorflow
Dataset
okk in this case I combined and saved all the images .png images as one file .npz file.
#for training data
datatrain = np.load('/kaggle/input/data-making-final/maps_256.npz')
X_train, y_train = datatrain['arr_0'], datatrain['arr_1']#for validation data
datavalid = np.load('/kaggle/input/data-making-final/Vmaps_256.npz')
X_val, y_val = datavalid['arr_0']*255, np.nan_to_num(datavalid['arr_1'])*255
#make sure there shapes are same
assert X_train.shape == y_train.shape
assert X_val.shape == y_val.shapeUNet model
Here the code for UNet model:
from tensorflow.keras import backend as K
from tensorflow.keras.layers import Input, Conv2D, MaxPooling2D, Conv2DTranspose, concatenate
from tensorflow.keras.models import Model
# Define the Dice coefficient metric for evaluating segmentation performance
def dice_coef(y_true, y_pred):
# Flatten the arrays to compute the intersection
y_true_f = K.flatten(y_true)
y_pred_f = K.flatten(y_pred)
intersection = K.sum(y_true_f * y_pred_f)
# Compute Dice coefficient
return (2. * intersection + 1) / (K.sum(y_true_f) + K.sum(y_pred_f) + 1)
# Define the loss function based on the Dice coefficient
def dice_coef_loss(y_true, y_pred):
# Negative Dice coefficient for use as a loss function
return -dice_coef(y_true, y_pred)
# Function to build the U-Net model
def unet(input_size=(256,256,1)):
# Input layer of the U-Net model
inputs = Input(input_size)
# Block 1: Convolution + pooling
conv1 = Conv2D(32, (3, 3), activation='relu', padding='same')(inputs)
conv1 = Conv2D(32, (3, 3), activation='relu', padding='same')(conv1)
pool1 = MaxPooling2D(pool_size=(2, 2))(conv1) # Pooling to reduce spatial dimensions
# Block 2: Convolution + pooling
conv2 = Conv2D(64, (3, 3), activation='relu', padding='same')(pool1)
conv2 = Conv2D(64, (3, 3), activation='relu', padding='same')(conv2)
pool2 = MaxPooling2D(pool_size=(2, 2))(conv2)
# Block 3: Convolution + pooling
conv3 = Conv2D(128, (3, 3), activation='relu', padding='same')(pool2)
conv3 = Conv2D(128, (3, 3), activation='relu', padding='same')(conv3)
pool3 = MaxPooling2D(pool_size=(2, 2))(conv3)
# Block 4: Convolution + pooling
conv4 = Conv2D(256, (3, 3), activation='relu', padding='same')(pool3)
conv4 = Conv2D(256, (3, 3), activation='relu', padding='same')(conv4)
pool4 = MaxPooling2D(pool_size=(2, 2))(conv4)
# Block 5: Convolution (without pooling)
conv5 = Conv2D(512, (3, 3), activation='relu', padding='same')(pool4)
conv5 = Conv2D(512, (3, 3), activation='relu', padding='same')(conv5)
# Block 6: Upsampling + concatenation + convolution
up6 = concatenate([Conv2DTranspose(256, (2, 2), strides=(2, 2), padding='same')(conv5), conv4], axis=3)
conv6 = Conv2D(256, (3, 3), activation='relu', padding='same')(up6)
conv6 = Conv2D(256, (3, 3), activation='relu', padding='same')(conv6)
# Block 7: Upsampling + concatenation + convolution
up7 = concatenate([Conv2DTranspose(128, (2, 2), strides=(2, 2), padding='same')(conv6), conv3], axis=3)
conv7 = Conv2D(128, (3, 3), activation='relu', padding='same')(up7)
conv7 = Conv2D(128, (3, 3), activation='relu', padding='same')(conv7)
# Block 8: Upsampling + concatenation + convolution
up8 = concatenate([Conv2DTranspose(64, (2, 2), strides=(2, 2), padding='same')(conv7), conv2], axis=3)
conv8 = Conv2D(64, (3, 3), activation='relu', padding='same')(up8)
conv8 = Conv2D(64, (3, 3), activation='relu', padding='same')(conv8)
# Block 9: Upsampling + concatenation + convolution
up9 = concatenate([Conv2DTranspose(32, (2, 2), strides=(2, 2), padding='same')(conv8), conv1], axis=3)
conv9 = Conv2D(32, (3, 3), activation='relu', padding='same')(up9)
conv9 = Conv2D(32, (3, 3), activation='relu', padding='same')(conv9)
# Output layer: 1x1 convolution to map the 32 feature channels to 1 output channel for binary segmentation
conv10 = Conv2D(1, (1, 1), activation='sigmoid')(conv9)
# Model construction
model = Model(inputs=inputs, outputs=conv10)
return model
model = unet(input_size=(256,256,3))This code below callbacks for a neural network model using TensorFlow’s Keras API. It includes ModelCheckpoint to save the best model weights based on validation loss, ReduceLROnPlateau to adjust learning rate when validation loss plateaus, and EarlyStopping to halt training if validation loss does not improve for a certain number of epochs. These callbacks aim to improve model performance and prevent overfitting during training.
from tensorflow.keras.callbacks import ModelCheckpoint, LearningRateScheduler, EarlyStopping, ReduceLROnPlateau
weight_path="{}_weights.best.hdf5".format('cxr_reg')
checkpoint = ModelCheckpoint(weight_path, monitor='val_loss', verbose=1,
save_best_only=True, mode='min', save_weights_only = True)
reduceLROnPlat = ReduceLROnPlateau(monitor='val_loss', factor=0.05, patience=3,
verbose=1, mode='min', epsilon=0.05, cooldown=2, min_lr=1e-6)
early = EarlyStopping(monitor="val_loss", mode="min", patience=15) # probably needs to be more patient, but kaggle time is limited
callbacks_list = [checkpoint, early, reduceLROnPlat]This code below sets up the compilation of a neural network model using TensorFlow’s Keras API. It uses the Adam optimizer with a learning rate of 0.001 and specifies the loss function as `dice_coef_loss`. Additionally, it defines metrics to monitor during training, including `dice_coef`, `binary_accuracy`, and `AUC` (Area Under the Curve).
Furthermore, it splits the dataset into training and validation sets using `train_test_split` from scikit-learn. The images are divided by 255 to normalize pixel values, and the mask is converted to binary format where values greater than 127 are considered as 1 and others as 0.
from IPython.display import clear_output
from tensorflow.keras.optimizers import Adam
from sklearn.model_selection import train_test_split
from sklearn.metrics import roc_curve, auc
model.compile(optimizer=Adam(learning_rate=0.001), loss=[dice_coef_loss], metrics = [dice_coef, 'binary_accuracy',"AUC" ])
#images, mask = images/255, (mask>127).astype(np.float32)
train_vol, validation_vol, train_seg, validation_seg = train_test_split((images)/255,
(mask>127).astype(np.float32),
test_size = 0.1)Now fit the model.
loss_history = model.fit(x = train_vol,y = train_seg,batch_size = 8,epochs = 100,validation_data =(validation_vol,validation_seg) , callbacks=callbacks_list)Please check the kaggle notebook here.
https://www.kaggle.com/code/sumitai/unet-on-data
If you enjoyed this article, I’d appreciate your applause, shares, and a follow — it’s a great encouragement for me to create more content :)
References




