Image Segmentation by UNet: Everything you need to know about Implementing in PyTorch
Image segmentation is a process in computer vision where an image is divided into different parts, with each part representing a specific…

Image segmentation is a process in computer vision where an image is divided into different parts, with each part representing a specific object or region. This technique helps a computer to identify and understand individual elements within an image, like distinguishing a cat from its background.
U-Net, a special type of neural network, excels in this task by analyzing an image at multiple scales, efficiently identifying and outlining intricate details and structures, thus improving the accuracy and precision of the segmentation process. This makes U-Net particularly effective in complex scenarios like medical image analysis, where precise differentiation of tissues or anomalies is crucial.
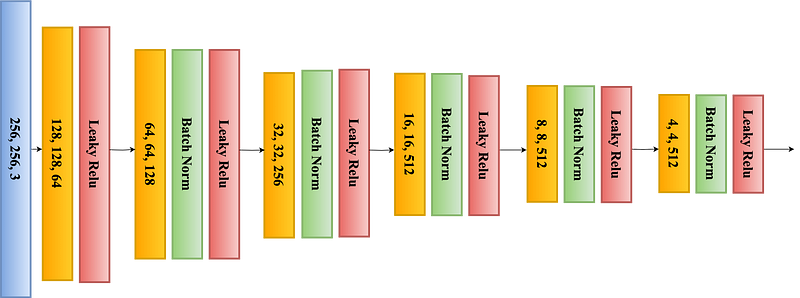
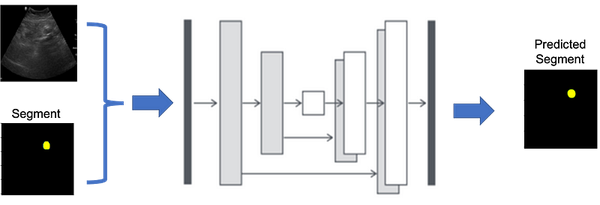
This structure, resembling the letter ‘U’, hence the name U-Net, allows for the network to efficiently learn from a limited amount of data. As shown in Figure 1, the contracting path is a typical CNN that captures features and reduces the spatial dimension, while the expanding path uses transposed convolutions to enable precise localization (as shown in Figure 2).
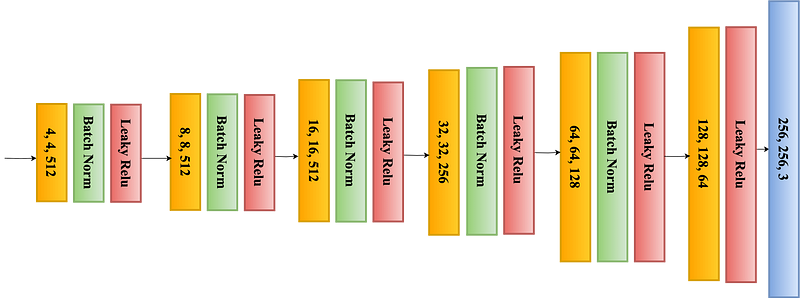
As compare to its predecessor models, U-Net’s capability to work with fewer training images while delivering highly accurate segmentations makes it ideal for medical imaging applications, where high-quality, annotated datasets are often scarce. Its success in medical imaging has also led to adaptations in various other domains requiring detailed image segmentation. Combining both encoder and decoder makes full UNet as shown in Figure 3 and combining

Here is plan of attack:
1. Import Libraries
2. Dataset
3. Transformation
4. UNet Blocks
5. UNet Model
6. Training Loop
7. Visualize the testing dataset
8. Save model
Import Libraries
import os
import glob
import random
import cv2
from PIL import Image
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
import torch
import torchvision
import torch.nn as nn
import torch.nn.functional as F
import torch.utils.data as data
from torch.utils.data import DataLoader
from mpl_toolkits.axes_grid1 import ImageGrid
from torchvision.utils import make_grid
import torchvision.transforms as tt
import albumentations as A
from sklearn.model_selection import train_test_split
from tqdm import tqdmcheck if GPU is available:
device = "cuda" if torch.cuda.is_available() else "cpu"
print("Using {} device".format(device))This code defines a `set_seed` function to ensure reproducibility in machine learning experiments by fixing random number generation with the given seed value. It sets the seed for PyTorch’s CPU and GPU operations, as well as for NumPy and Python’s random module, all with the same specified seed value (defaulting to 0).
def set_seed(seed = 0):
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
torch.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
np.random.seed(seed)
random.seed(seed)
set_seed()Dataset
This code prepares a DataFrame `files_df` containing image and mask file paths, along with a diagnosis label. It first identifies mask files using glob, derives image file paths from them, and then assigns a diagnosis label based on whether the maximum pixel value in the mask is greater than 0 (1 if true, 0 otherwise). The resulting DataFrame contains columns “image_path,” “mask_path,” and “diagnosis,” summarizing this information.
ROOT_PATH = '../input/lgg-mri-segmentation/kaggle_3m/'
mask_files = glob.glob(ROOT_PATH + '*/*_mask*')
image_files = [file.replace('_mask', '') for file in mask_files]
def diagnosis(mask_path):
return 1 if np.max(cv2.imread(mask_path)) > 0 else 0
files_df = pd.DataFrame({"image_path": image_files,
"mask_path": mask_files,
"diagnosis": [diagnosis(x) for x in mask_files]})
files_dfThis code creates a stacked bar plot to visualize the data distribution of images with and without tumors. It sets the plot’s title, labels, and x-axis tick labels. Additionally, it annotates the bar segments with the respective counts for “No Tumor” and “Tumor” categories, enhancing the readability of the plot.
ax = files_df['diagnosis'].value_counts().plot(kind='bar', stacked=True, figsize=(6,6), color=['green', 'red'])
ax.set_title('Data Distribution', fontsize=15)
ax.set_ylabel('No. of Images', fontsize=15)
ax.set_xticklabels(['No Tumor', 'Tumor'], fontsize=12, rotation=0)
for i, rows in enumerate(files_df['diagnosis'].value_counts().values):
ax.annotate(int(rows), xy=(i, rows+12), ha='center', fontweight='bold', fontsize=12)This code splits the `files_df` DataFrame into three sets: training, validation, and test sets. It first splits the data into a training set and a validation set with a 90–10 split ratio, stratifying based on the “diagnosis” column to ensure class balance. Then, it further splits the training set into a new training set and a test set with a 85–15 split ratio. The resulting data set shapes are printed to provide an overview of the data split.
- Training set shape: (85% of original data)
- Validation set shape: (10% of original data)
- Test set shape: (5% of original data)
train_df, val_df = train_test_split(files_df, stratify=files_df['diagnosis'], test_size=0.1, random_state=0)
train_df = train_df.reset_index(drop=True)
val_df = val_df.reset_index(drop=True)
train_df, test_df = train_test_split(train_df, stratify=train_df['diagnosis'], test_size=0.15, random_state=0)
train_df = train_df.reset_index(drop=True)
test_df = test_df.reset_index(drop=True)
print("Train: {}\nVal: {}\nTest: {}".format(train_df.shape, val_df.shape, test_df.shape))This code snippet generates a visual representation of brain MRI images and their corresponding masks. It selects five random samples from the training data with a diagnosis of 1 (indicating the presence of a tumor), loads their images and masks, and displays them using Matplotlib’s ImageGrid. The resulting grid contains three subplots:
- The first subplot displays the raw brain MRI images.
- The second subplot shows the corresponding masks.
- The third subplot overlays the masks on top of the MRI images, allowing visualization of the region affected by the tumor.
This visualization is useful for inspecting and understanding the data used in the machine learning experiment.
set_seed()
images, masks = [], []
df_positive = train_df[train_df['diagnosis']==1].sample(5).values
for sample in df_positive:
img = cv2.imread(sample[0])
mask = cv2.imread(sample[1])
images.append(img)
masks.append(mask)
images = np.hstack(np.array(images))
masks = np.hstack(np.array(masks))
fig = plt.figure(figsize=(15,10))
grid = ImageGrid(fig, 111, nrows_ncols=(3,1), axes_pad=0.4)
grid[0].imshow(images)
grid[0].set_title('Images', fontsize=15)
grid[0].axis('off')
grid[1].imshow(masks)
grid[1].set_title('Masks', fontsize=15)
grid[1].axis('off')
grid[2].imshow(images)
grid[2].imshow(masks, alpha=0.4)
grid[2].set_title('Brain MRI with mask', fontsize=15)
grid[2].axis('off')This code defines a custom dataset class called `BrainDataset` for working with brain MRI images and their corresponding masks. It’s designed to be used with PyTorch’s `DataLoader` for training and evaluation. Here’s a breakdown of what the class does:
- The constructor `__init__` initializes the dataset with a DataFrame `df` containing image and mask file paths and an optional `transform` argument for data augmentation.
- `__len__` returns the total number of samples in the dataset, which is the length of the DataFrame.
- `__getitem__` loads an image and its mask at the specified index `idx`. It reads the image and mask using OpenCV, scales their values to the range [0, 1], and applies the provided transformations if any.
- The image is then preprocessed, including transposing the channels from HWC to CHW format, converting it to a PyTorch tensor of type `torch.float32`, and normalizing it using mean and standard deviation values.
- The mask is similarly preprocessed, expanded to have a single channel, transposed, and converted to a PyTorch tensor of type `torch.float32`.
- Finally, it returns the preprocessed image and mask as a tuple.
This dataset class can be used to create data loaders for training and evaluation in a deep learning model, allowing for easy handling of the data during training.
class BrainDataset(data.Dataset):
def __init__(self, df, transform=None):
self.df = df
self.transform = transform
def __len__(self):
return len(self.df)
def __getitem__(self, idx):
image = cv2.imread(self.df.iloc[idx, 0])
image = np.array(image)/255.
mask = cv2.imread(self.df.iloc[idx, 1], 0)
mask = np.array(mask)/255.
if self.transform is not None:
aug = self.transform(image=image, mask=mask)
image = aug['image']
mask = aug['mask']
image = image.transpose((2,0,1))
image = torch.from_numpy(image).type(torch.float32)
image = tt.Normalize((0.485, 0.456, 0.406), (0.229, 0.224, 0.225))(image)
mask = np.expand_dims(mask, axis=-1).transpose((2,0,1))
mask = torch.from_numpy(mask).type(torch.float32)
return image, maskTransformation
These are the data augmentation transformations defined using the `albumentations` library for the training, validation, and test sets:
- `train_transform`:
- Resizes the image to a width and height of 128 pixels.
- Applies horizontal flipping with a 50% probability.
- Applies vertical flipping with a 50% probability.
- Randomly rotates the image by 90 degrees with a 50% probability.
- Shifts, scales, and rotates the image with specified limits and probabilities.
2. `val_transform`:
- Resizes the image to a width and height of 128 pixels.
- Applies horizontal flipping with a 50% probability.
3. `test_transform`:
- Resizes the image to a width and height of 128 pixels.
These transformations are commonly used for data augmentation in computer vision tasks, such as image segmentation, to increase the diversity of the training data and improve the model’s generalization. During training, you would apply `train_transform`, and during validation and testing, you would apply `val_transform` and `test_transform`, respectively, to preprocess the images consistently.
train_transform = A.Compose([
A.Resize(width=128, height=128, p=1.0),
A.HorizontalFlip(p=0.5),
A.VerticalFlip(p=0.5),
A.RandomRotate90(p=0.5),
A.ShiftScaleRotate(shift_limit=0.01, scale_limit=0.04, rotate_limit=0, p=0.25),
])
val_transform = A.Compose([
A.Resize(width=128, height=128, p=1.0),
A.HorizontalFlip(p=0.5),
])
test_transform = A.Compose([
A.Resize(width=128, height=128, p=1.0)
])set_seed()
train_ds = BrainDataset(train_df, train_transform)
val_ds = BrainDataset(val_df, val_transform)
test_ds = BrainDataset(test_df, test_transform)The dataset_info function provides information about a given dataset, including its size and the size of a random sample from it
def dataset_info(dataset):
print(f'Size of dataset: {len(dataset)}')
index = random.randint(1, 40)
img, label = dataset[index]
print(f'Sample-{index} Image size: {img.shape}, Mask: {label.shape}\n')
print('Train dataset:')
dataset_info(train_ds)
print('Validation dataset:')
dataset_info(val_ds)
print('Test dataset:')
dataset_info(test_ds)These code snippets create data loaders for the training, validation, and test datasets using PyTorch’s `DataLoader` class. Here’s a breakdown of each:
- `train_dl`:
- Data loader for the training dataset (`train_ds`).
- `batch_size`: The number of samples per batch is set to 64.
- `shuffle=True`: The data is shuffled before each epoch during training.
- `num_workers=2`: Two worker processes are used to load data in parallel.
- `pin_memory=True`: Pinning memory for faster GPU data transfer, suitable for systems with GPU acceleration.
Same steps goes for val_dl and test_dl
These data loaders are typically used for training and evaluating machine learning models in PyTorch.
batch_size = 64
set_seed()
train_dl = DataLoader(train_ds,
batch_size,
shuffle=True,
num_workers=2,
pin_memory=True)
set_seed()
val_dl = DataLoader(val_ds,
batch_size,
num_workers=2,
pin_memory=True)
test_dl = DataLoader(val_ds,
batch_size,
num_workers=2,
pin_memory=True)
images, masks = next(iter(train_dl))
print(images.shape)
print(masks.shape)The code defines two functions and uses them to visualize a batch of images and masks from the training data loader (`train_dl`). Here’s what each function does:
- `denormalize (images)`: This function denormalizes the input images by reversing the normalization applied during data preprocessing. It uses the means and standard deviations of the ImageNet dataset to perform the denormalization.
- `show_batch (dl)`: This function takes a data loader (`dl`) as input and iterates through a batch of data from it. For each batch:
- It creates two subplots. The first subplot (`fig1`) displays the denormalized images, while the second subplot (`fig2`) displays the masks.
- The images are denormalized using the `denormalize` function and displayed as a grid using `make_grid`.
- The masks are displayed in a similar manner.
- The loop breaks after processing the first batch, so only one batch of data is visualized.
Overall, this code snippet is useful for visualizing a batch of images and their corresponding masks from the training data loader, allowing you to inspect the data and ensure that preprocessing and denormalization are correctly applied.
def denormalize(images):
means = torch.tensor([0.485, 0.456, 0.406]).reshape(1, 3, 1, 1)
stds = torch.tensor([0.229, 0.224, 0.225]).reshape(1, 3, 1, 1)
return images * stds + means
def show_batch(dl):
for images, masks in dl:
fig1, ax1 = plt.subplots(figsize=(24, 24))
ax1.set_xticks([]); ax1.set_yticks([])
denorm_images = denormalize(images)
ax1.imshow(make_grid(denorm_images[:13], nrow=13).permute(1, 2, 0).clamp(0,1))
fig2, ax2 = plt.subplots(figsize=(24, 24))
ax2.set_xticks([]); ax2.set_yticks([])
ax2.imshow(make_grid(masks[:13], nrow=13).permute(1, 2, 0).clamp(0,1))
break
show_batch(train_dl)UNet Blocks
The code defines several essential components for building a U-Net-like architecture for image segmentation using PyTorch:
- `DoubleConv`: This class represents a double convolution block, consisting of two sequential convolutional layers, each followed by batch normalization and ReLU activation.
- `Down`: This class represents the downsampling portion of the network, which combines max-pooling with the `DoubleConv` block for feature extraction.
- `Up`: This class represents the upsampling portion of the network. It can use either bilinear upsampling or transposed convolution (depending on the `bilinear` flag) to increase the spatial resolution of the input. It then concatenates the upsampled feature maps with the corresponding feature maps from the contracting path before applying the `DoubleConv` block.
- `OutConv`: This class represents the final output convolutional layer with a sigmoid activation, typically used to produce segmentation masks.
These components are building blocks for constructing a U-Net architecture, which is a popular choice for image segmentation tasks. You can use these modules to create the U-Net model by defining the architecture in terms of the down, up, and output layers, and then connecting them accordingly.
class DoubleConv(nn.Module):
"""(convolution => [BN] => ReLU) * 2"""
def __init__(self, in_channels, out_channels, mid_channels=None):
super().__init__()
if not mid_channels:
mid_channels = out_channels
self.double_conv = nn.Sequential(
nn.Conv2d(in_channels, mid_channels, kernel_size=3, padding=1),
nn.BatchNorm2d(mid_channels),
nn.ReLU(inplace=True),
nn.Conv2d(mid_channels, out_channels, kernel_size=3, padding=1),
nn.BatchNorm2d(out_channels),
nn.ReLU(inplace=True))
def forward(self, x):
return self.double_conv(x)
class Down(nn.Module):
"""Downscaling with maxpool then double conv"""
def __init__(self, in_channels, out_channels):
super().__init__()
self.maxpool_conv = nn.Sequential(
nn.MaxPool2d(2),
DoubleConv(in_channels, out_channels))
def forward(self, x):
return self.maxpool_conv(x)
class Up(nn.Module):
"""Upscaling then double conv"""
def __init__(self, in_channels, out_channels, bilinear=True):
super().__init__()
if bilinear:
self.up = nn.Upsample(scale_factor=2, mode='bilinear', align_corners=True)
self.conv = DoubleConv(in_channels, out_channels, in_channels//2)
else:
self.up = nn.ConvTranspose2d(in_channels, in_channels//2, kernel_size=2, stride=2)
self.conv = DoubleConv(in_channels, out_channels)
def forward(self, x1, x2):
x1 = self.up(x1)
diffY = x2.size()[2] - x1.size()[2]
diffX = x2.size()[3] - x1.size()[3]
x1 = F.pad(x1, [diffX//2, diffX-diffX//2,
diffY//2, diffY-diffY//2])
x = torch.cat([x2, x1], dim=1)
return self.conv(x)
class OutConv(nn.Module):
def __init__(self, in_channels, out_channels):
super(OutConv, self).__init__()
self.conv = nn.Sequential(
nn.Conv2d(in_channels, out_channels, kernel_size=1),
nn.Sigmoid())
def forward(self, x):
return self.conv(x)UNet Model
The `UNet` class defines a U-Net architecture for image segmentation using PyTorch. Here’s a breakdown of the components and the architecture:
- `n_channels`: The number of input channels (e.g., 3 for RGB images).
- `n_classes`: The number of output classes (e.g., 1 for binary segmentation).
- `bilinear`: A boolean flag indicating whether to use bilinear interpolation during upsampling.
The architecture consists of the following components:
- `inc`: The initial convolutional layer, followed by a double convolution block.
- `down1`, `down2`, `down3`, and `down4`: Downscaling blocks that reduce spatial dimensions and increase the number of feature maps.
- `up1`, `up2`, `up3`, and `up4`: Upscaling blocks that upsample feature maps and concatenate them with corresponding feature maps from the contracting path.
- `outc`: The final output convolutional layer with a sigmoid activation.
The `forward` method defines the forward pass of the U-Net model. It sequentially passes the input through the encoding and decoding paths and returns the logits (unnormalized predictions) for each pixel in the segmentation mask.
This U-Net architecture is commonly used for various image segmentation tasks, including medical image segmentation and general-purpose semantic segmentation. You can create an instance of this `UNet` class with the desired number of input channels and output classes to build and train your image segmentation model.
class UNet(nn.Module):
def __init__(self, n_channels, n_classes, bilinear=True):
super(UNet, self).__init__()
self.n_channels = n_channels
self.n_classes = n_classes
self.bilinear = bilinear
self.inc = DoubleConv(n_channels, 64)
self.down1 = Down(64, 128)
self.down2 = Down(128, 256)
self.down3 = Down(256, 512)
factor = 2 if bilinear else 1
self.down4 = Down(512, 1024//factor)
self.up1 = Up(1024, 512//factor, bilinear)
self.up2 = Up(512, 256//factor, bilinear)
self.up3 = Up(256, 128//factor, bilinear)
self.up4 = Up(128, 64, bilinear)
self.outc = OutConv(64, n_classes)
def forward(self, x):
x1 = self.inc(x)
x2 = self.down1(x1)
x3 = self.down2(x2)
x4 = self.down3(x3)
x5 = self.down4(x4)
x = self.up1(x5, x4)
x = self.up2(x, x3)
x = self.up3(x, x2)
x = self.up4(x, x1)
logits = self.outc(x)
return logitsIn this code, you have created an instance of the `UNet` model with 3 input channels and 1 output channel (typically used for binary segmentation). You have also moved the model to the specified `device`. Then, you have passed a random input tensor of shape (1, 3, 128, 128) to the model and printed the shape of the output tensor.
The output shape `out.shape` should be (1, 1, 128, 128), indicating that the model has produced a segmentation mask with a single channel (1) for the 128x128 input image. Each value in the output tensor represents the model’s prediction for the corresponding pixel in the segmentation mask.
model = UNet(3, 1).to(device)
out = model(torch.randn(1, 3, 128, 128).to(device))
print(out.shape)These are three functions used for training and evaluating a deep learning model for image segmentation, particularly for binary segmentation tasks. Here’s an overview of each function:
- `dice_coef_metric(pred, label)`: This function calculates the Dice coefficient, which is a common evaluation metric for image segmentation tasks. It measures the similarity between the predicted segmentation mask (`pred`) and the ground truth mask (`label`). The Dice coefficient is defined as `(2 * intersection) / (union)`, where `intersection` is the element-wise multiplication of the prediction and label masks, and `union` is the sum of their values. It returns a value between 0 and 1, where a higher value indicates better segmentation performance.
- `dice_coef_loss(pred, label)`: This function computes the Dice coefficient loss. It’s used as a loss function during training to encourage the model to produce segmentation masks that are similar to the ground truth masks. It adds a small smoothing term to the numerator and denominator to prevent division by zero and returns `1 — (Dice coefficient)` as the loss.
- `bce_dice_loss(pred, label)`: This function combines two loss components: the binary cross-entropy (BCE) loss (`bce_loss`) and the Dice coefficient loss (`dice_loss`). The BCE loss measures the pixel-wise binary classification loss, while the Dice loss encourages segmentation mask similarity. By combining these losses, you aim to strike a balance between pixel-level accuracy and mask similarity during training. This combined loss is often used as the final loss function when training a model for binary image segmentation.
You can use these functions to evaluate segmentation performance using the Dice coefficient and train your model with the combined `bce_dice_loss` as the loss function.
def dice_coef_metric(pred, label):
intersection = 2.0 * (pred * label).sum()
union = pred.sum() + label.sum()
if pred.sum() == 0 and label.sum() == 0:
return 1.
return intersection / union
def dice_coef_loss(pred, label):
smooth = 1.0
intersection = 2.0 * (pred * label).sum() + smooth
union = pred.sum() + label.sum() + smooth
return 1 - (intersection / union)
def bce_dice_loss(pred, label):
dice_loss = dice_coef_loss(pred, label)
bce_loss = nn.BCELoss()(pred, label)
return dice_loss + bce_lossTraining loop
This is a training loop for training a deep learning model for image segmentation. It’s designed to train the model using a given data loader (`loader`) and a specified loss function (`loss_func`). Here’s an explanation of what the code does:
- `model.train()`: Puts the model in training mode, enabling gradient computation and dropout layers if present.
- `train_losses` and `train_dices`: Lists to store the training losses and Dice coefficients for each batch.
- The loop iterates over the batches from the data loader (`loader`), where each batch contains input images and their corresponding ground truth masks (`image` and `mask`).
- For each batch, the following steps are performed:
- The input images and masks are moved to the specified device (e.g., GPU) for computation.
- The model forward pass is executed, producing predicted segmentation masks (`outputs`)
- A thresholding operation is applied to `outputs` to convert them into binary masks (`out_cut`). Values less than 0.5 are set to 0, and values greater than or equal to 0.5 are set to 1. This is often done to convert model predictions into clear binary masks.
- The Dice coefficient is calculated using the `dice_coef_metric` function to evaluate the model’s segmentation performance for the current batch.
- The loss is computed using the specified loss function (`loss_func`) by comparing the predicted masks (`outputs`) to the ground truth masks (`mask`).
- The loss is appended to the `train_losses` list, and the Dice coefficient is appended to the `train_dices` list.
- After processing all batches, the gradients are backpropagated (`loss.backward()`), and the optimizer’s parameters are updated (`optimizer.step()`). Gradients are then zeroed to prepare for the next batch (`optimizer.zero_grad()`).
- The function returns `train_dices` and `train_losses`, which can be used to monitor the training progress and evaluate the model’s performance over epochs.
This training loop is suitable for binary image segmentation tasks, and it computes both the loss and Dice coefficient as metrics for training and evaluation.
def train_loop(model, loader, loss_func):
model.train()
train_losses = []
train_dices = []
# for i, (image, mask) in enumerate(tqdm(loader)):
for i, (image, mask) in enumerate(loader):
image = image.to(device)
mask = mask.to(device)
outputs = model(image)
out_cut = np.copy(outputs.data.cpu().numpy())
out_cut[np.nonzero(out_cut < 0.5)] = 0.0
out_cut[np.nonzero(out_cut >= 0.5)] = 1.0
dice = dice_coef_metric(out_cut, mask.data.cpu().numpy())
loss = loss_func(outputs, mask)
train_losses.append(loss.item())
train_dices.append(dice)
loss.backward()
optimizer.step()
optimizer.zero_grad()
return train_dices, train_lossesThis code defines a function `eval_loop` that evaluates a model using a given dataset loader and loss function. It is designed to work in both evaluation and training modes, controlled by the `training` parameter. Here’s a brief explanation of the key steps:
1. Set Model to Evaluation Mode: `model.eval()` switches the model to evaluation mode, affecting layers like dropout and batch normalization which behave differently during training and evaluation.
2. Initialize Metrics: Variables `val_loss` and `val_dice` are initialized to accumulate loss and Dice coefficient (a metric for segmentation tasks) values over the dataset.
3. Loop Over Dataset: The loop iterates over the dataset provided by `loader`, where each iteration provides a batch of `image` and `mask` pairs. The `image` is the input to the model, and `mask` is the true segmentation map for evaluating the model’s predictions.
4. Data to Device: Both `image` and `mask` are moved to the computation device (like a GPU), using `.to(device)`.
5. Model Prediction: `model(image)` generates the predicted segmentation maps for the input images.
6. Calculate Loss: The `loss_func` computes the loss between the model’s predictions and the true masks.
7. Thresholding Predictions: The predictions (`outputs`) are thresholded at 0.5 to convert probabilities into binary segmentation maps, with values below 0.5 set to 0 and values equal or above 0.5 set to 1.
8. Dice Coefficient Calculation: `dice_coef_metric` calculates the Dice coefficient between the thresholded predictions and the true masks, which is a measure of the overlap between the two.
9. Accumulate Metrics: Both loss and Dice coefficient for each batch are accumulated to compute the average over the entire dataset.
10. Average Metrics: The total accumulated `val_loss` and `val_dice` are divided by the number of steps (batches) to calculate the mean loss and Dice coefficient over the dataset.
11. Scheduler Step in Training Mode: If the function is called in training mode (`training=True`), it adjusts the learning rate based on the mean Dice coefficient using a learning rate scheduler (`scheduler.step(val_mean_dice)`).
12. Return Metrics: The function returns the average Dice coefficient and loss across the dataset.
This function is useful for evaluating the performance of a segmentation model on a validation set during training or for testing the model’s performance on a test set.
def eval_loop(model, loader, loss_func, training=True):
model.eval()
val_loss = 0
val_dice = 0
with torch.no_grad():
for step, (image, mask) in enumerate(loader):
image = image.to(device)
mask = mask.to(device)
outputs = model(image)
loss = loss_func(outputs, mask)
out_cut = np.copy(outputs.data.cpu().numpy())
out_cut[np.nonzero(out_cut < 0.5)] = 0.0
out_cut[np.nonzero(out_cut >= 0.5)] = 1.0
dice = dice_coef_metric(out_cut, mask.data.cpu().numpy())
val_loss += loss
val_dice += dice
val_mean_dice = val_dice / step
val_mean_loss = val_loss / step
if training:
scheduler.step(val_mean_dice)
return val_mean_dice, val_mean_lossThis code defines a function `train_model` to train a segmentation model using training and validation data loaders, a specified loss function, an optimizer, a scheduler, and a number of epochs. Here’s a breakdown of the process:
1. Initialize History Lists: Four lists are initialized to keep track of training and validation loss and Dice scores over each epoch.
2. Epoch Loop: The training process is iterated over a specified number of epochs. An epoch is one full pass through the entire training dataset.
3. Training Phase:
- `train_loop` function is called with the model, training data loader, and loss function, returning lists of Dice coefficients and losses for each batch in the training set.
- The mean training Dice coefficient and loss are calculated by averaging the Dice coefficients and losses over all training batches.
4. Validation Phase:
- `eval_loop` function is used to evaluate the model on the validation dataset with the same loss function. It returns the mean Dice coefficient and loss for the validation set.
- Unlike in the `train_loop`, the model parameters are not updated during the `eval_loop`.
5. Record History: The mean training and validation losses and Dice coefficients for the current epoch are appended to their respective history lists.
6. Logging: At the end of each epoch, the mean losses and Dice coefficients for both training and validation phases are printed out. This provides insight into how the model is performing and whether it is improving, overfitting, or underfitting.
7. Return Histories: After completing all epochs, the function returns the histories of training and validation losses and Dice coefficients. These can be used for analyzing the model’s performance over time, such as plotting learning curves.
This function encapsulates the entire training process, including forward passes to compute losses, backward passes to compute gradients, parameter updates, and model evaluation on a validation set to monitor performance. The scheduler, which adjusts the learning rate based on certain criteria (not explicitly shown in this snippet), is presumably used within the `train_loop`.
def train_model(train_loader, val_loader, loss_func, optimizer, scheduler, num_epochs):
train_loss_history = []
train_dice_history = []
val_loss_history = []
val_dice_history = []
for epoch in range(num_epochs):
train_dices, train_losses = train_loop(model, train_loader, loss_func)
train_mean_dice = np.array(train_dices).mean()
train_mean_loss = np.array(train_losses).mean()
val_mean_dice, val_mean_loss = eval_loop(model, val_loader, loss_func)
train_loss_history.append(np.array(train_losses).mean())
train_dice_history.append(np.array(train_dices).mean())
val_loss_history.append(val_mean_loss)
val_dice_history.append(val_mean_dice)
print('Epoch: {}/{} | Train Loss: {:.3f}, Val Loss: {:.3f}, Train DICE: {:.3f}, Val DICE: {:.3f}'.format(epoch+1, num_epochs,
train_mean_loss,
val_mean_loss,
train_mean_dice,
val_mean_dice))
return train_loss_history, train_dice_history, val_loss_history, val_dice_historyoptimizer = torch.optim.Adam(model.parameters(), lr=1e-3)
scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(optimizer, 'max', patience=3)
num_epochs = 30
%%time
train_loss_history, train_dice_history, val_loss_history, val_dice_history = train_model(train_dl, val_dl, bce_dice_loss, optimizer, scheduler, num_epochs)Visualize the testing dataset
Rest of these codes are just plotting the results.
def plot_dice_history(model_name, train_dice_history, val_dice_history, num_epochs):
x = np.arange(num_epochs)
fig = plt.figure(figsize=(10, 6))
plt.plot(x, train_dice_history, label='Train DICE', lw=3, c="b")
plt.plot(x, val_dice_history, label='Validation DICE', lw=3, c="r")
plt.title(f"{model_name}", fontsize=20)
plt.legend(fontsize=12)
plt.xlabel("Epoch", fontsize=15)
plt.ylabel("DICE", fontsize=15)
plt.show()
plot_dice_history('UNET', train_dice_history, val_dice_history, num_epochs)def plot_loss_history(model_name, train_loss_history, val_loss_history, num_epochs):
x = np.arange(num_epochs)
fig = plt.figure(figsize=(10, 6))
plt.plot(x, train_loss_history, label='Train Loss', lw=3, c="b")
plt.plot(x, val_loss_history, label='Validation Loss', lw=3, c="r")
plt.title(f"{model_name}", fontsize=20)
plt.legend(fontsize=12)
plt.xlabel("Epoch", fontsize=15)
plt.ylabel("Loss", fontsize=15)
plt.show()
plot_loss_history('UNET', train_loss_history, val_loss_history, num_epochs)%%time
test_dice, test_loss = eval_loop(model, test_dl, bce_dice_loss, training=False)
print("Mean IoU/DICE: {:.3f}%, Loss: {:.3f}".format((100*test_dice), test_loss))test_sample = test_df[test_df["diagnosis"] == 1].sample(24).values[0]
image = cv2.resize(cv2.imread(test_sample[0]), (128, 128))
mask = cv2.resize(cv2.imread(test_sample[1]), (128, 128))
# pred
pred = torch.tensor(image.astype(np.float32) / 255.).unsqueeze(0).permute(0,3,1,2)
pred = tt.Normalize((0.485, 0.456, 0.406), (0.229, 0.224, 0.225))(pred)
pred = model(pred.to(device))
pred = pred.detach().cpu().numpy()[0,0,:,:]
pred_t = np.copy(pred)
pred_t[np.nonzero(pred_t < 0.3)] = 0.0
pred_t[np.nonzero(pred_t >= 0.3)] = 255.
pred_t = pred_t.astype("uint8")
# plot
fig, ax = plt.subplots(nrows=2, ncols=2, figsize=(10, 10))
ax[0, 0].imshow(image)
ax[0, 0].set_title("image")
ax[0, 1].imshow(mask)
ax[0, 1].set_title("mask")
ax[1, 0].imshow(pred)
ax[1, 0].set_title("prediction")
ax[1, 1].imshow(pred_t)
ax[1, 1].set_title("prediction with threshold")
plt.show()Save Model
Finally we can save the model just by using this command ‘torch.save’
torch.save(model.state_dict(), 'brain-mri-unet.pth')So here is everything you need to know about Segmentation by UNet I hope you like this. Happy Learning :)
Reference:
U-Net: Convolutional Networks for Biomedical Image Segmentation https://arxiv.org/abs/1505.04597